研究活動 (Sk Istiaque AHMED)
Sampling influences on detection of fish species by MiFish analysis
Environmental DNA (eDNA) is gaining immense popularity in understanding fish species distribution pattern and biodiversity monitoring in the ocean due to its non-invasive nature and convenience to use.
The eDNA is derived from the genetic materials present in water and gleaned from various sources including skin cells, feces and mucous.
MiFish metabarcoding technique is a modern technique in analyzing fish community distribution in larger waterbodies which capacitates detection of huge number of species at a short time.
Despite its easiness in use and application, MiFish metabarcoding technique isn’t devoid of demerits that include several challenges such as amplification failure, dominance of non-target taxa and lack of variation in the amplified fragments.
To circumvent these challenges, the current study buckles down to decide on appropriate sampling process between intake, bucket and niskin samples for MiFish analysis.
|
1. Study site Samples around Japan. 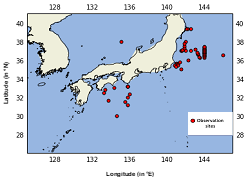 |
2. Sample collection Sampling waters by Niskin bottles, bucket, and intake. 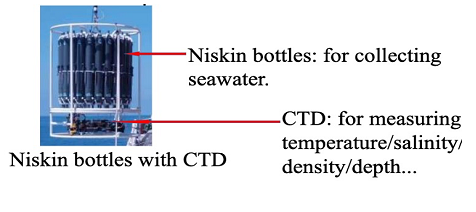 |
3. Filtering Water filteration by using pump and Sterivex filter.  |
|
4. Storing Addition of RNAlater and storing the samples at -30 degC.  |
5. eDNA extraction Extract environmental DNA (eDNA) in open ocean: OceanDNA.  |
6. MiFish analysis MiFish analysis (Miya et al., 2020) to identify fish species. The MiFish Metabarcoding result shows a highly diversified habitat of the study site as about 250 fish species from 100 families have been identified. |
|
7. Data processing Data processing and cluster analysis by using Ward method and Euclidean distance. Samples from Niskin bottle, bucket, and intake were compared. The detection percentage of most of the species did not vary significantly among different sampling types while some species vary significantly among each sampling type. |
8. Principal results For frequently detected species, most of them did not show any significant difference in the detection percentage. 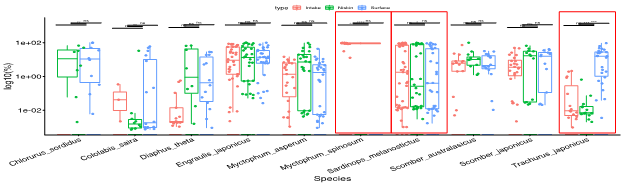 |
9. Future analysis The results support an idea that the Niskin, intake and surface water samples can be used as a mixture or simultaneously for MiFish analysis. We will further investigate the influence of environments and species dependency for the detection difference between the three types of sampling. |
